Types of Labelling for Microscopy
The types of labelling used depends on the type of microscope or imaging technique.
1. Transmission Electron Microscope (TEM), Scanning Electron Microscope (SEM), Serial Block Face-SEM, etc. requires using heavy metals such as Osmium to label tissues.
2. Light Microscopes are used to image samples labelled based on Immunocytochemistry (ICC), Immunohistochemistry (IHC) and Immunofluorescence (IF), all of which utilize antibodies to provide visual details about protein abundance, distribution, and localization.
Labelling For Light Microscopes
What samples are used in ICC vs IHC?
Sample Type and Preparation - tissue vs cells – As the name implies, IHC detection refers to tissue immunostaining, of either formalin fixed paraffin-embedded (FFPE) or frozen tissue. ICC refers to the staining of isolated or cultured intact cells where samples may be from tissue culture cell lines, either adherent or in suspension.
Immunocytochemistry (ICC)
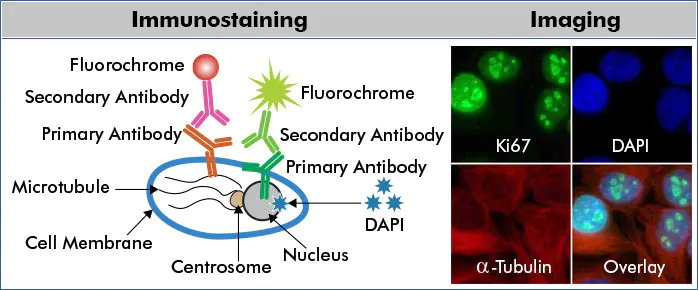
Immunocytochemistry (ICC) is an immunostaining technique that detects antigens in cultured or primary cells via light microscopy. Fluorochromes are the most common reporter molecules in microscopy and are standardly used in multicolor ICC experiments.
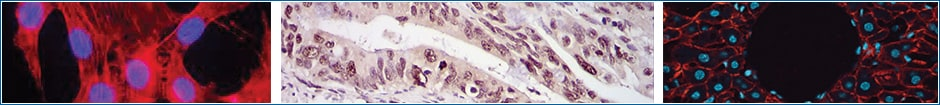
The example above depicts a multicolor ICC/IF scheme for indirect protein detection. HeLa cells were probed with anti-Ki67 (nuclear protein) and anti-Alpha-tubulin (microtubular protein) followed by detection with DyLightTM488 (green) and DyLightTM550 (red) conjugated secondary antibodies, respectively. Fixed cells were counterstained with the nuclear dye, DAPI (blue)
Immunohistochemistry (IHC)
Immunohistochemistry (IHC) is a technique that uses antibodies conjugated to enzymes that catalyze reactions to form detectable compounds to visualize and localize specific antigens in a tissue sample. The root “histo-” specifically applies to biological tissue, so the process is only immunohistochemistry if it is being done in organic tissue. In contrast, immunocytochemistry applies the same process to individual cells.
First, the tissue and cells must be “fixed” using a chemical like formaldehyde. This stabilizes the structural properties of cells, preventing them from changing throughout the process. Next, cells need to be permeated using a detergent such as Triton X, which allows antibodies to enter the tissue and bind to epitopes within the cell.
Primary antibodies against a protein of interest are added, and secondary antibodies with enzymes like horseradish peroxidase (HRP) conjugated to their Fc domain are added to target the primary antibody. Enzymes like HRP can target certain substrate molecules like diaminobenzidine (DAB) and catalyze oxidation which results in the creation of a colorful compound.
This colorful compound will stay localized to the area the antibody was targeted to, staining the area near the protein of interest a different color from the rest of the tissue. Lastly, tissues are counter-stained using a dye like a hematoxylin to create contrast between the tissue stained using IHC and the non-colored regions for better visualization.
Immunohistochemistry (IHC) has found numerous applications in medicine, especially in cancer diagnosis. Lymphomas are among the cancers most dependent on IHC for correct diagnosis and treatment decisions.
Direct vs indirect immunofluorescence (IF)
Immunofluorescence (IF) or cell imaging techniques rely on the use antibodies to label a specific target antigen with a fluorescent dye (also called fluorophores or fluorochromes) such as fluorescein isothiocyanate (FITC). Primary conjugated antibodies that are chemically labeled to fluorophores are commonly used in IF.
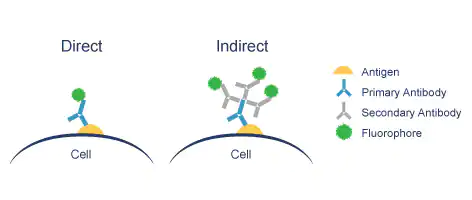
We distinguish between two IF methods depending on whether the fluorophore is conjugated to the primary or the secondary antibody:
Direct IF uses a single antibody directed against the target of interest. The primary antibody is directly conjugated to a fluorophore.
Indirect IF uses two antibodies. The primary antibody is unconjugated and a fluorophore-conjugated secondary antibody directed against the primary antibody is used for detection.
Labelling For Super-Resolution Microscopes
Super-resolution fluorescence microscopy, also called “nanoscopy,” enables the visualization of cellular structures beyond the diffraction limit of light. However, unlike electron microscopy, whose application is limited to fixed cells, nanoscopy enables live-cell imaging to study cellular dynamics in unprecedented spatial detail.
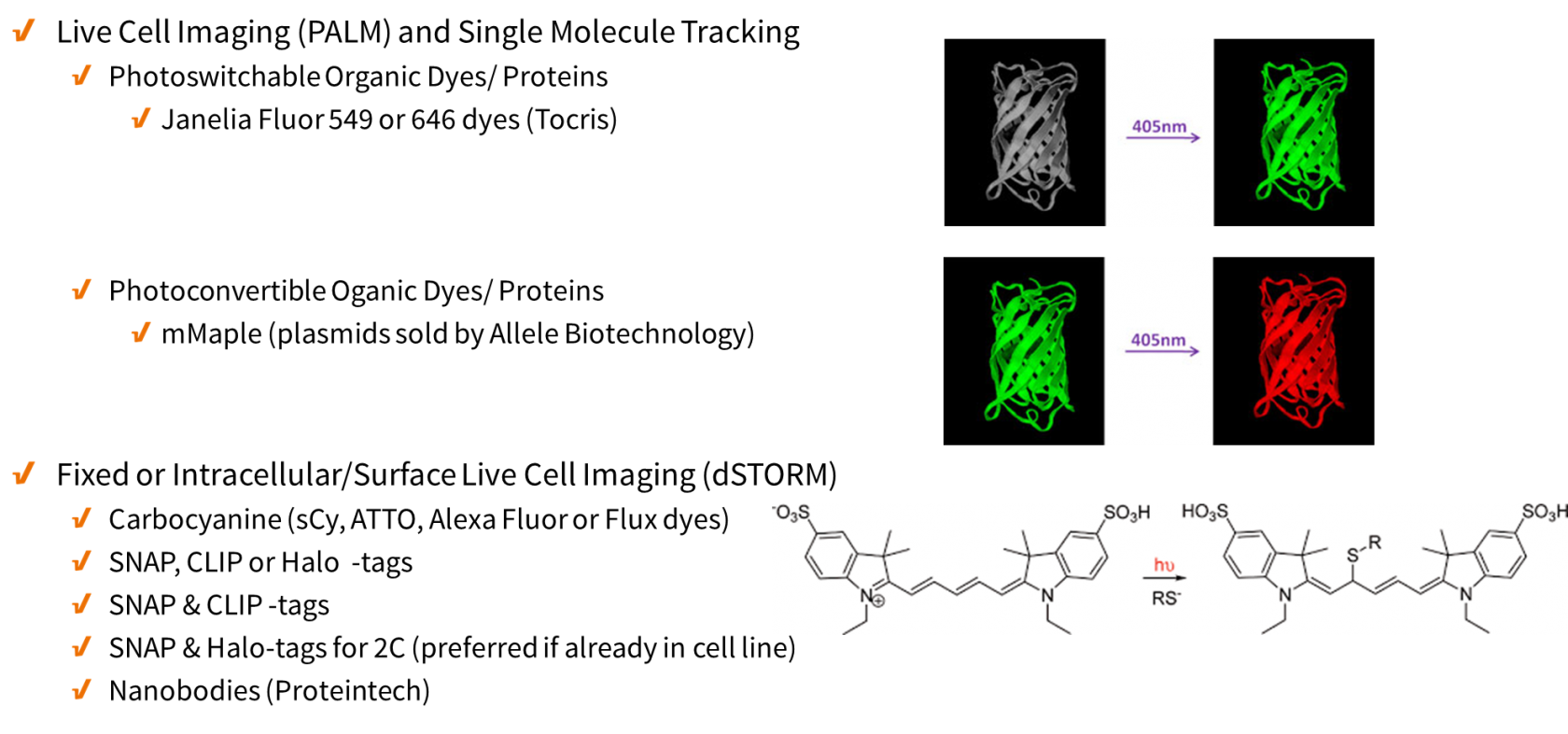
Green fluorescent protein (GFP) and its spectral variants have revolutionized biology, as they allow genetically encoded labeling, but they possess mediocre photophysical properties, generally emitting fewer photons than the best organic dyes by one or two orders of magnitude.
For super-resolution imaging, organic fluorophores are highly appealing because of their brightness and photostability . Organic fluorophores can be attached to proteins by combining click chemistry with unnatural amino acid incorporation. A second option is the direct coupling to proteins in live cells by using self-labeling proteins such as SNAP-tags (or a variant called CLIP-tag; and HaloTags. Alternatively, labeling can be achieved by combining click chemistry and self-labeling proteins. Like GFP, these self-labeling SNAP-tags and HaloTags can be expressed as fusion proteins and selectively reacted with the substrates benzylguanine (BG) and chloralkane (CA), respectively, which are tagged with organic fluorophores.
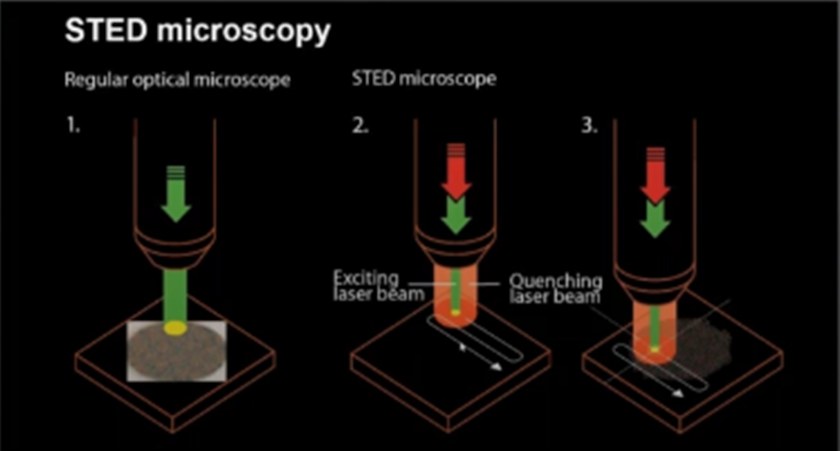
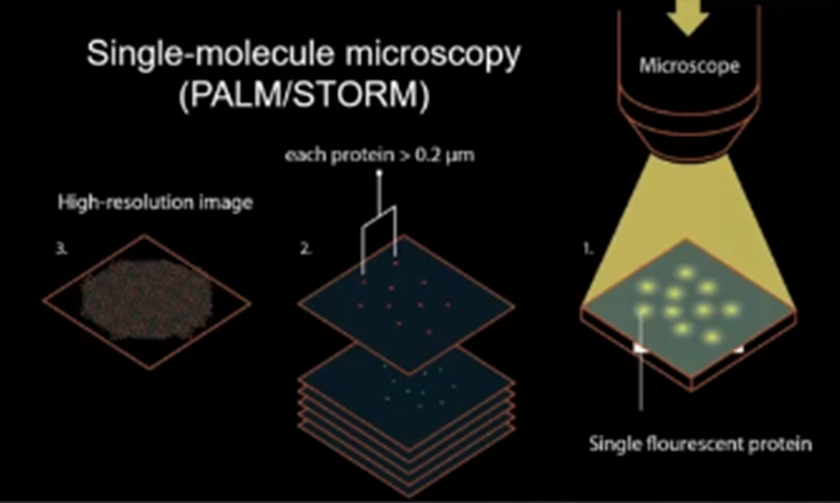
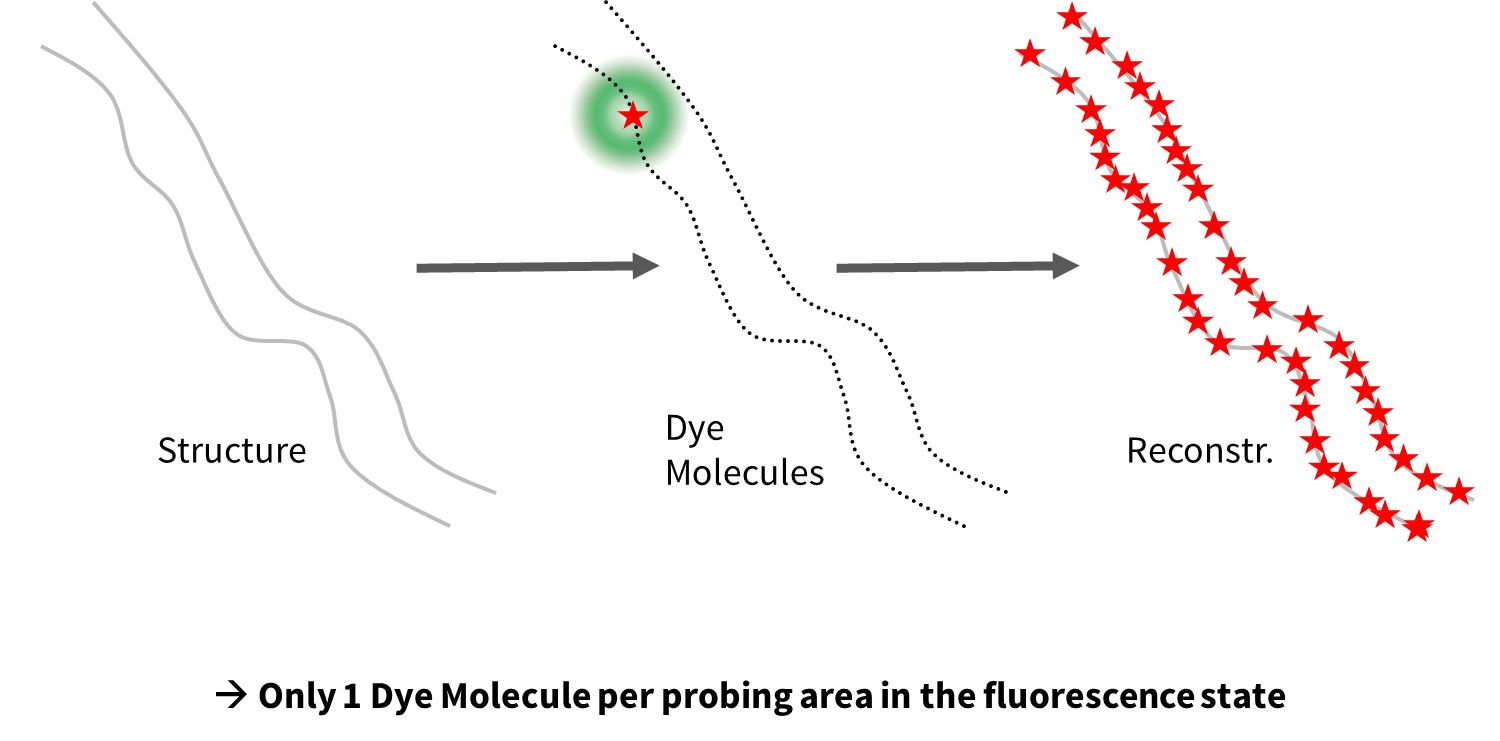
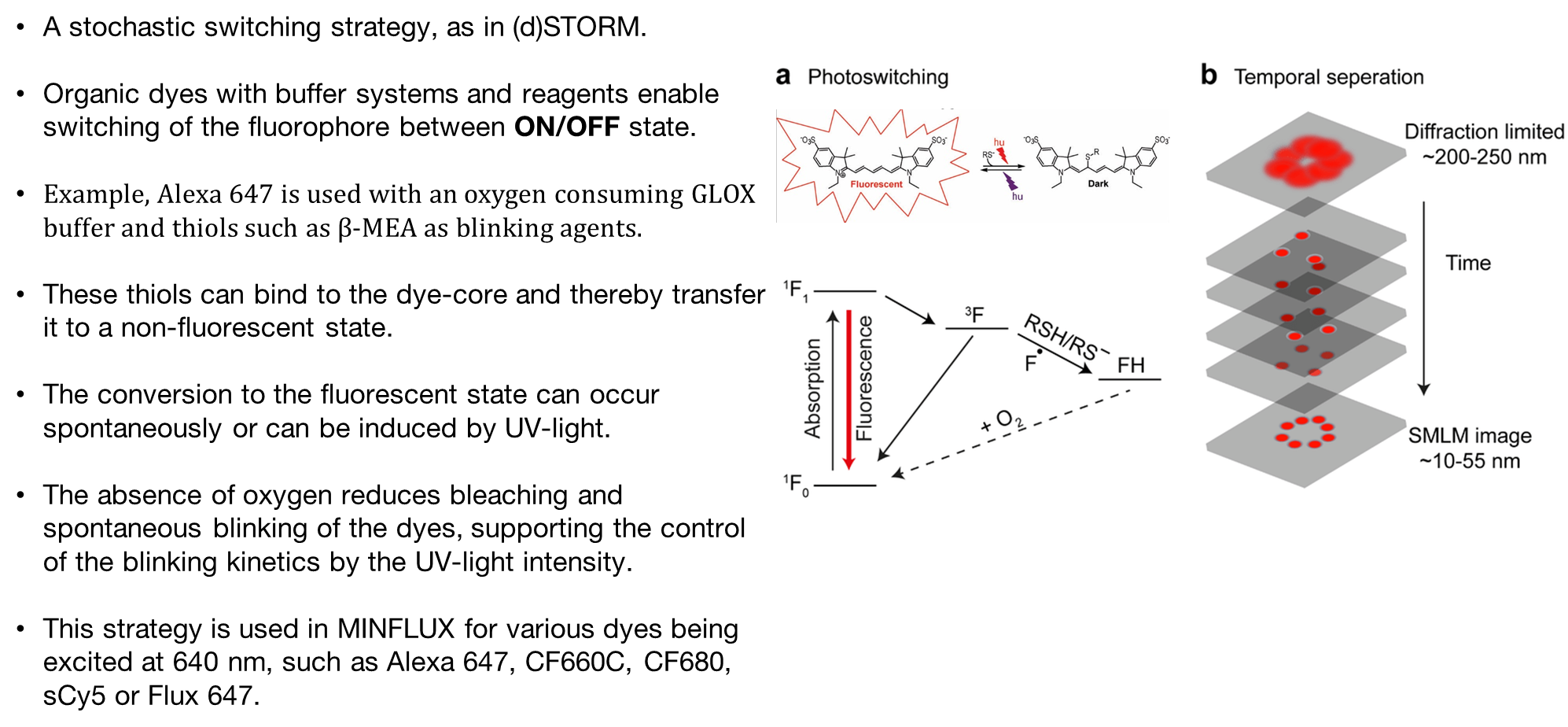
Super-resolution techniques such as STED, PALM, dSTORM and Minflux localizes one molecule at a time. Single molecule localization techniques use ON/OFF principle.
STED uses a beam that turns fluorescence on, and another beam, shaped like a “doughnut” that turns fluorescence off allowing single molecules to be detected as it scans across the sample.
PALM/STORM uses same on/off concept but turns molecules individually off and on using photoactivatable proteins
Minflux combines STED and dSTOM or PALM principles in imaging.
- Live Cell Imaging (similar to PALM/STORM) – no blinking buffer needed
- Fixed or Surface Marker Live Cell Imaging (similar to dSTORM) – may require blinking buffer
- Snap-tags, a self-labeling protein tag commercially available in various expression vectors that can be fused to any protein of interest and further specifically and covalently tagged with a suitable ligand, such as a fluorescent dye (snap-tag 647 is recommended).
- New England Biolabs sell substrates that label the SNAP-tag® without the need for additional enzymes. Cell-permeable substrates (SNAP-Cell®) are suitable for both intracellular and cell-surface labeling, whereas non-cell-permeable substrates (SNAP-Surface®) are specific for fusion proteins expressed on the cell surface only.
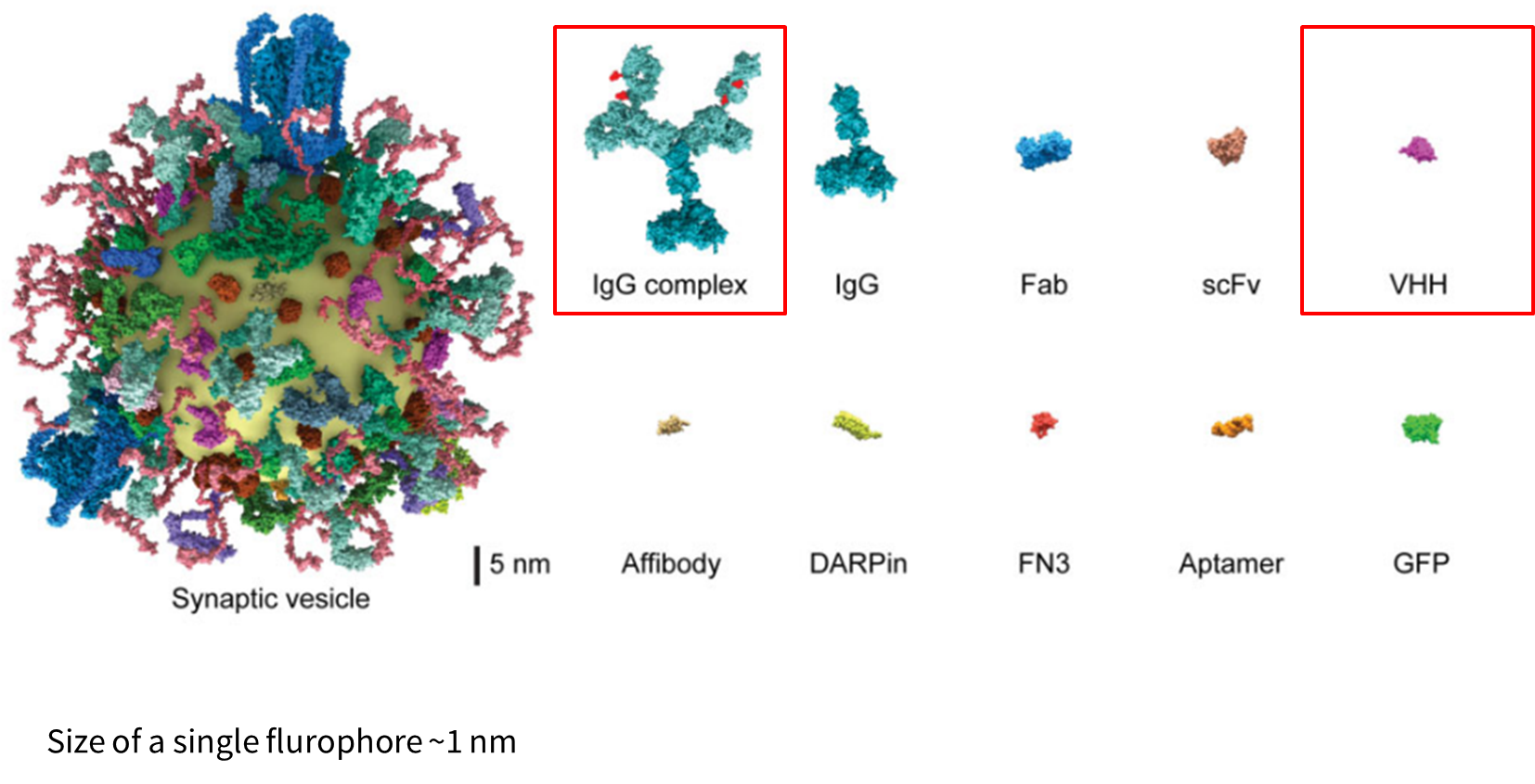
Size of Labels
- A single IgG is about 15nm and so the IgG complex could be >25nm
- In contrast, VHH nanobodies are ~3nm
- Proteintech sells ChromoTek SNAP/CLIP-tag VHH, recombinant binding protein
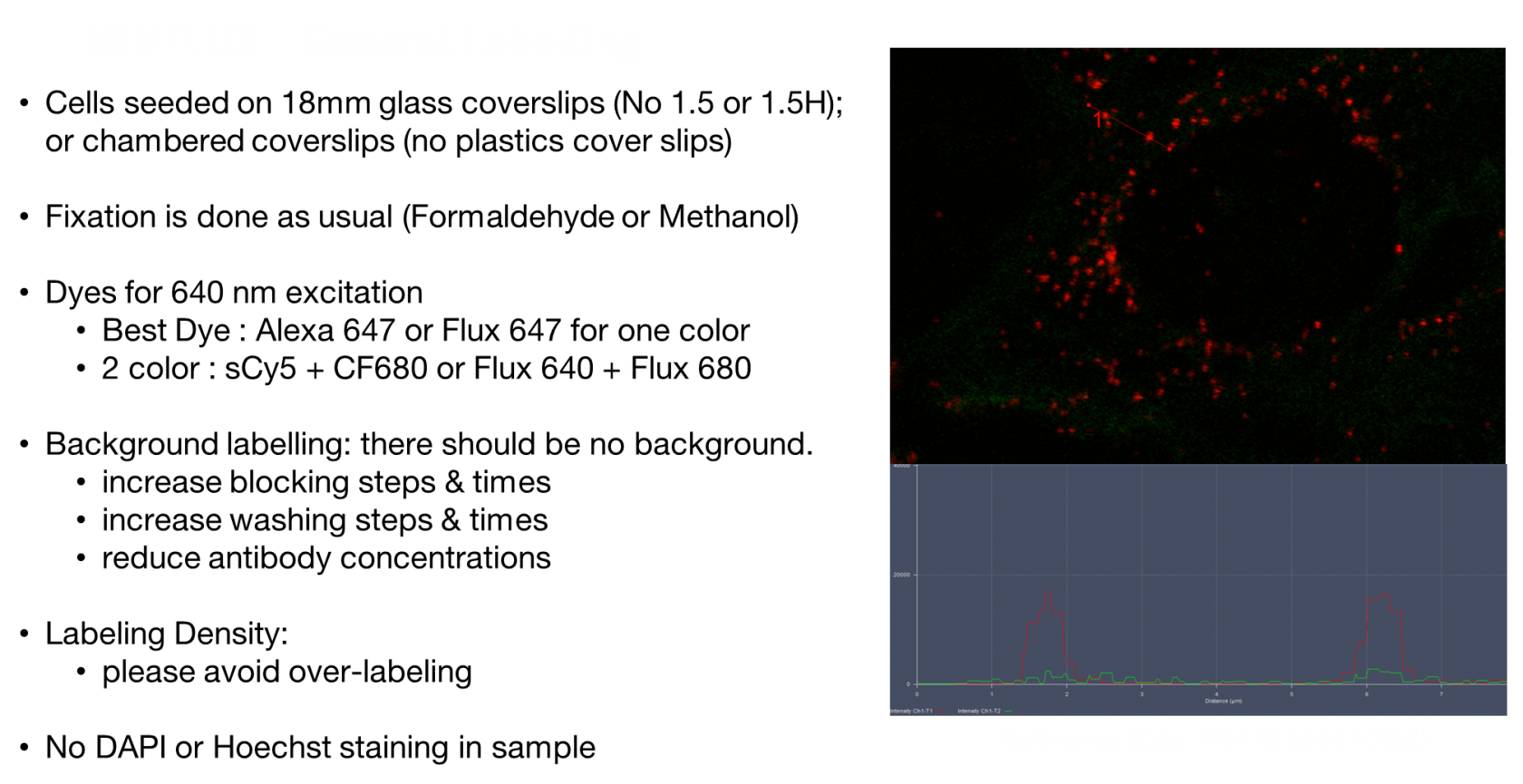
Minflux General Labelling